https://doi.org/10.1140/epjs/s11734-024-01347-4
Regular Article
High- accuracy chaotic time series prediction of the flexible beam-ring model based on P CNN-BiLSTM ED network
CNN-BiLSTM ED network
1
School of Applied Science, Beijing Information Science and Technology University, 100192, Beijing, China
2
College of Mechanical Engineering, Beijing Information Science and Technology University, 100192, Beijing, China
Received:
18
July
2024
Accepted:
16
September
2024
Published online:
9
October
2024
In this paper, data-driven modeling is used to predict the chaotic time series of a two-degree-of-freedom nonlinear system of the beam-ring model. To accurately predict the chaotic time series of the system, the long short-term memory (LSTM) neural network is improved under the framework of n + 1 neural networks for n-degree-of-freedom systems. First, a bidirectional encoder is introduced into the LSTM network to capture the dependencies within the data set of the system, and the prediction accuracy is improved by combining the information from both directions of the sequence. At the same time, the introduction of a convolutional neural network (CNN) can effectively extract the features of the input dataset and simplify the amount of data in the input encoder. Therefore, we propose a new neural network model that combines a convolutional neural network and long short-term memory based on a bidirectional encoder–decoder hybrid network (P CNN-BiLSTM ED). The results of numerical experiments show that the prediction accuracy of P
CNN-BiLSTM ED). The results of numerical experiments show that the prediction accuracy of P CNN-BiLSTM ED is significantly improved in each plane of the system. The accuracy of the plane
CNN-BiLSTM ED is significantly improved in each plane of the system. The accuracy of the plane 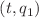 is increased by 65.49%. In parallel, the enhanced methodology (P
is increased by 65.49%. In parallel, the enhanced methodology (P CNN-BiLSTM ED) can be employed to predict chaotic time series of complex dynamic systems.
CNN-BiLSTM ED) can be employed to predict chaotic time series of complex dynamic systems.
Copyright comment Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
© The Author(s), under exclusive licence to EDP Sciences, Springer-Verlag GmbH Germany, part of Springer Nature 2024
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.




